Our research group focuses on developing computational approaches to analyze high-throughput drug and genetic screens, along with multi-omics datasets, to study the fundamental regulatory mechanisms underlying cancer cell response to treatments. Working closely with experimental and clinical groups, we develop integrative approaches to address one of the most pressing challenges in cancer: drug resistance.
We aim to develop the next generation of multidisciplinary engineers who can bridge the fields of computer science, cell biology, and biomedicine.
Machine Learning | Multi-omics | Functional Genomics | Drug resistance | Cancer
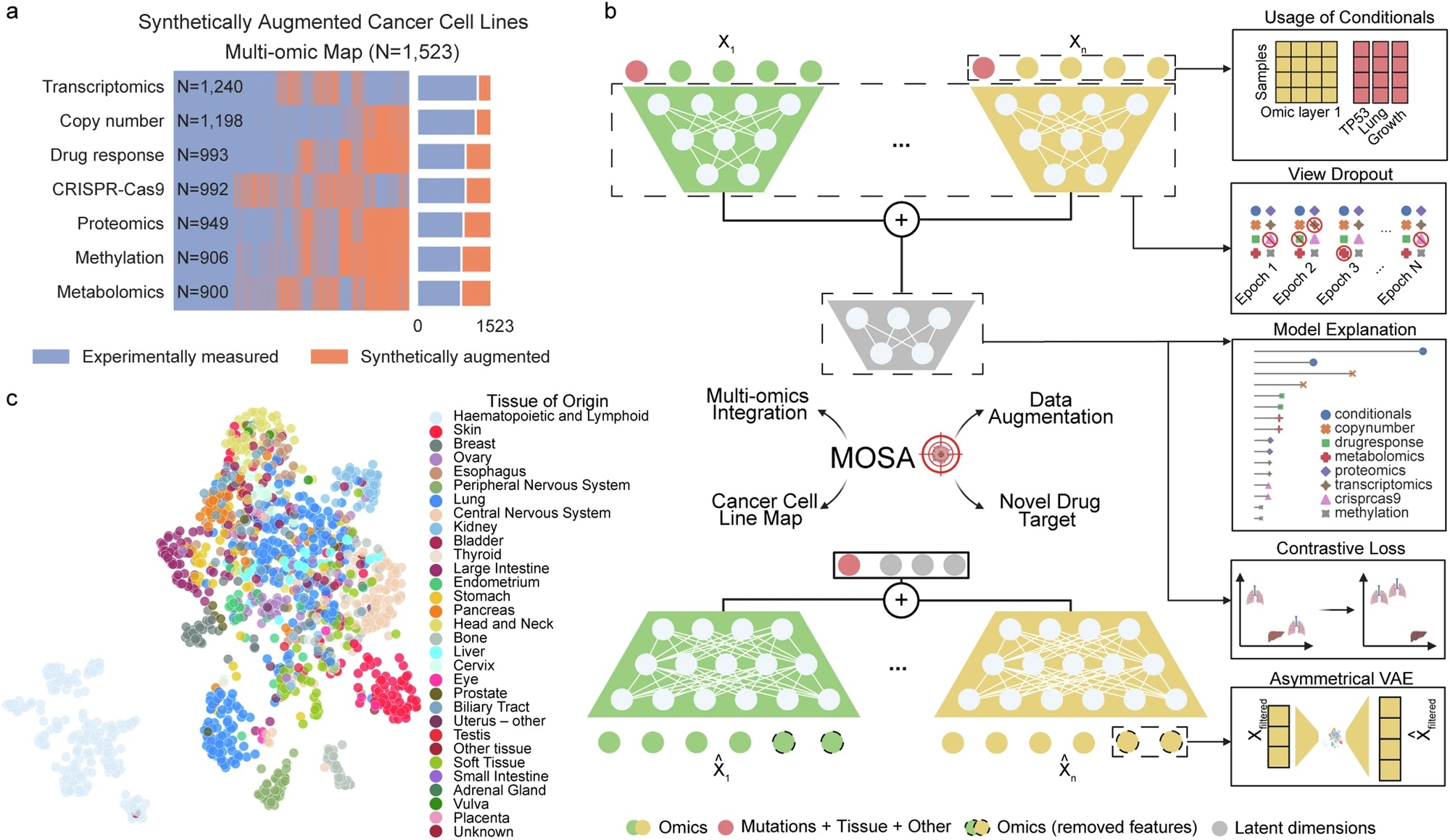
This study presents the first synthetic Cancer Dependency Map (DepMap) using a novel deep generative...
[Read More]
Fulbright Scholarship
Deep learning application to combinatorial saturation mutagenesis screens
Emanuel will visit the Broad Institute of MIT and Harvard, where we will work with...
[Read More]
Cancer Dependency Map 2.0
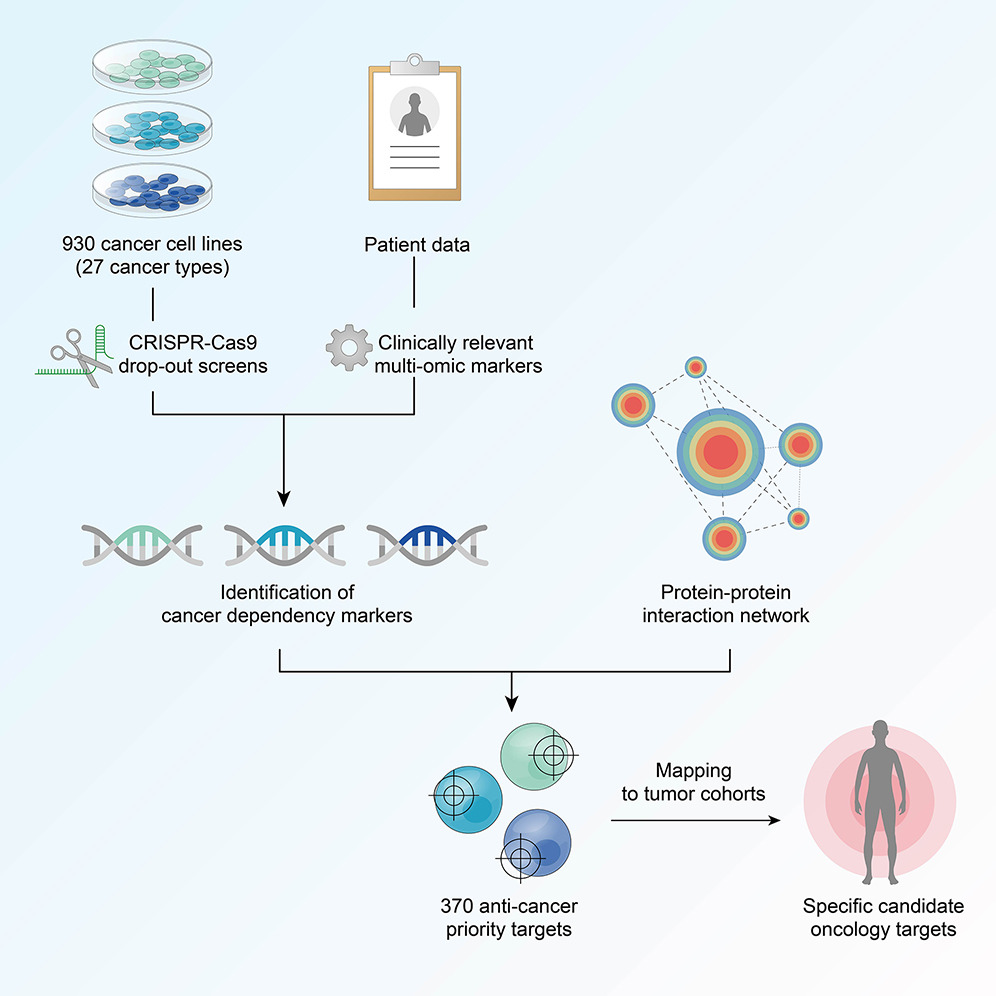
A comprehensive clinically informed map of dependencies in cancer cells and framework for target prioritization
In our study, we developed a second-generation map of cancer dependencies by annotating 930 cancer...
[Read More]
Base editing screens
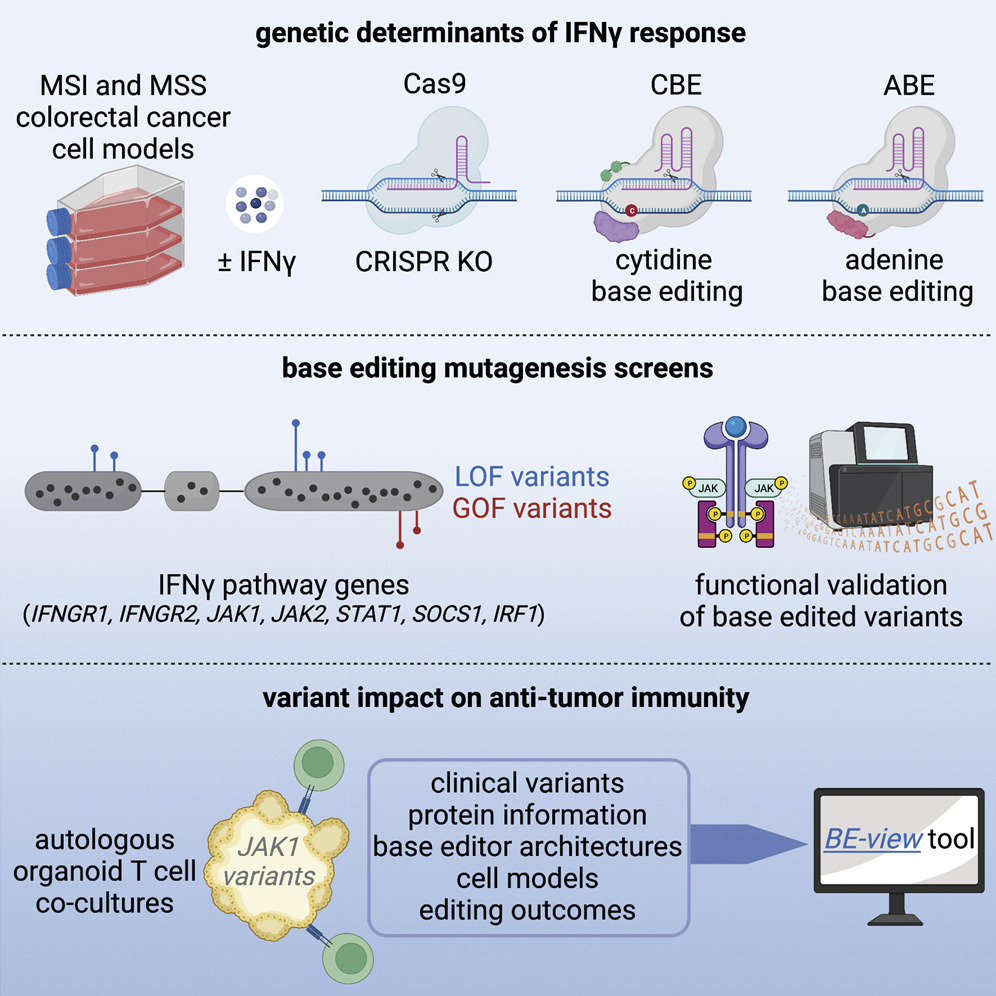
Base editing screens map mutations affecting interferon-gamma signaling in cancer
In this study, we introduce a new method for identifying the effects of genetic variations...
[Read More]
Efficient large-scale cancer organoid technique
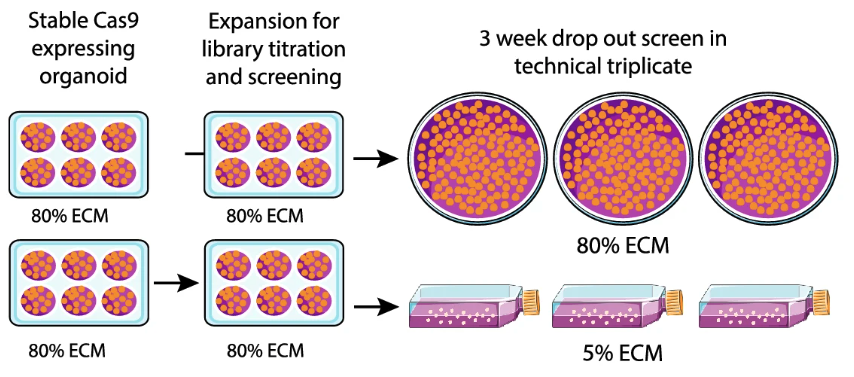
A suspension technique for efficient large-scale cancer organoid culturing and perturbation screens
High-throughput technique that allows functional screening, pharmacological and functional genomics, in cancer organoids.
[Read More]